Recently, I am diving into some deep learning for genomics. My go-to deep-learning framework Keras has a nice feature fit_generator that fetches mini-batches of data from a indefinite python generator and train the neural network incrementally using each mini-batch. The python generator should yield a batch of feature and label from every next(generator) call. An example is as follow:
class data_generator():
'''
suppose the input data file is two columns:
where first columns is feature, second column is label
'''
def __init__(self, tsv_file, batch_size = 500):
self.X = []
self.y = []
self.batch_size = batch_size
self.data_file = tsv_file
self.sample_generator = open(self.data_file)
def __next__(self):
# reinitiate samples
self.X = []
self.y = []
# populate this batch with features and labels
data_gen()
# return for keras model fit_generater
return np.array(self.X), np.array(self.y)
def data_gen(self):
sample_count = 0
while self.batch_size > sample_count: # break loop when batch is filled
try:
line = next(self.sample_generator)
feature, label = line.split('\t') ## extract feature and labels from the two columns
self.X.append(feature)
self.y.append(label)
sample_count += 1
except StopIteration: # if it loops to the end (finished one epoch), re-open the file and loop again
self.sample_generator = open(self.data_file) ## open the file again
line = next(self.sample_generator)
feature, label = line.split('\t') ## extract feature and labels from the two columns
self.X.append(feature)
self.y.append(label)
sample_count += 1
However, one drawback of this generator is that batches are created sequentially from the data file, such that training samples are not shuffled. To introduce randomness into mini-batches, we can add a line of if random.random() > 0.5: before putting sample into the batch:
def data_gen(self):
sample_count = 0
while self.batch_size > sample_count: # break loop when batch is filled
try:
line = next(self.sample_generator)
if random.random() > 0.5: ### added randomness ###
feature, label = line.split('\t') ## extract feature and labels from the two columns
self.X.append(feature)
self.y.append(label)
sample_count += 1
except StopIteration: # if it loops to the end, re-open the file and loop again
self.sample_generator = open(self.data_file) ## open the file again
line = next(self.sample_generator)
if random.random() > 0.5: ### added randomness ###
feature, label = line.split('\t') ## extract feature and labels from the two columns
self.X.append(feature)
self.y.append(label)
sample_count += 1
The builtin random module in python is nice enough to generator a number between 0 and 1, but it can be a bit slow. So in this post, I will show an implementation of random float number between 0 and 1 using cython and see how much speed up we can get.
Below is the cython random function:
%%cython
from libc.stdlib cimport rand, RAND_MAX
cpdef double cy_random():
return rand()/RAND_MAX
Let’s check if the results are similar by looking at the distibution of 10000 random numbers:
import random
import matplotlib.pyplot as plt
import seaborn as sns
ax = plt.subplot(111)
sns.distplot([random.random() for i in range(10000)], ax = ax, label='builtin')
sns.distplot([cy_random() for i in range(10000)], ax = ax, label = 'Cython')
ax.legend(fontsize=15, bbox_to_anchor = (1,0.5))
ax.set_xlabel('Random number', fontsize = 15)
ax.set_ylabel('Density', fontsize=15)
sns.despine()
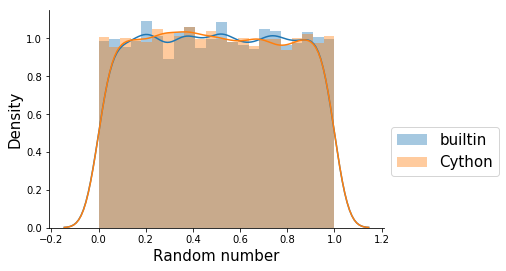
That looks similar enough, both of them are more or less uniform over [0,1]. Now, lets see how much time it takes for each of them to run:
%timeit random.random()
108 ns ± 7.83 ns per loop (mean ± std. dev. of 7 runs, 10000000 loops each)
%timeit cy_random()
54.2 ns ± 2.73 ns per loop (mean ± std. dev. of 7 runs, 10000000 loops each)
The cython version yielded a 2X speed-up.
