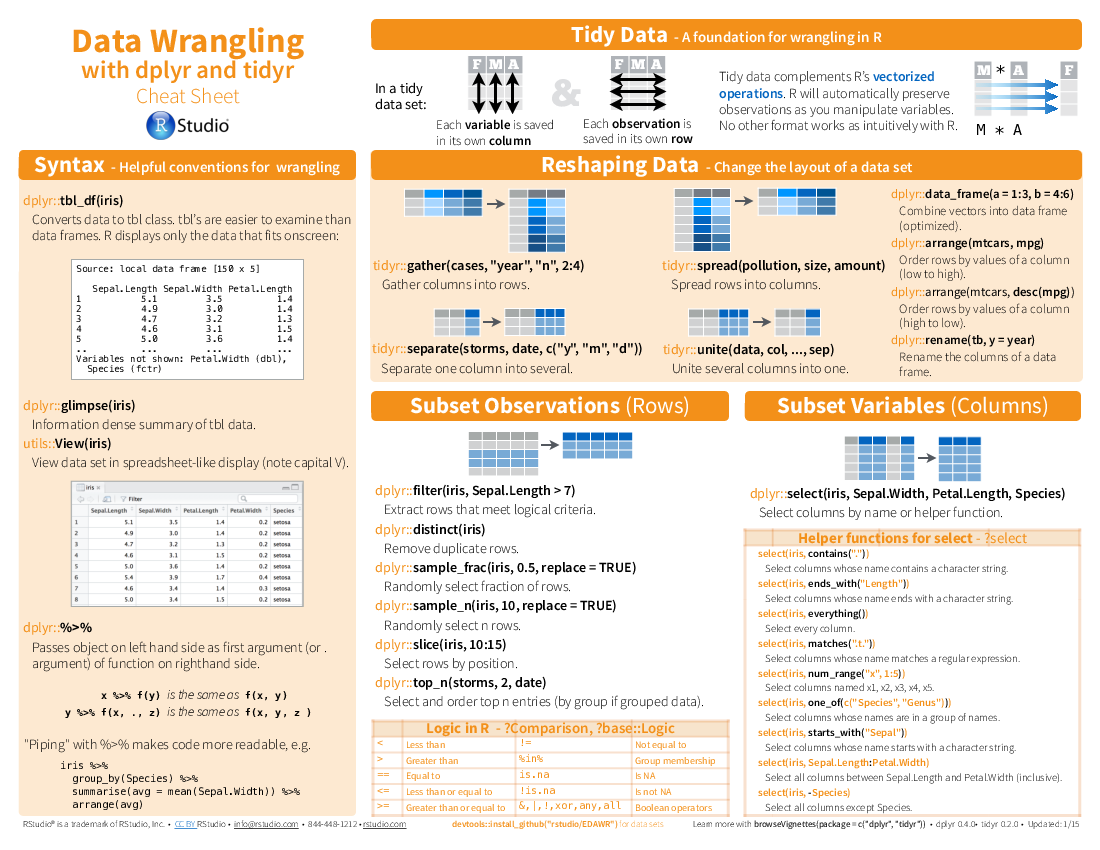
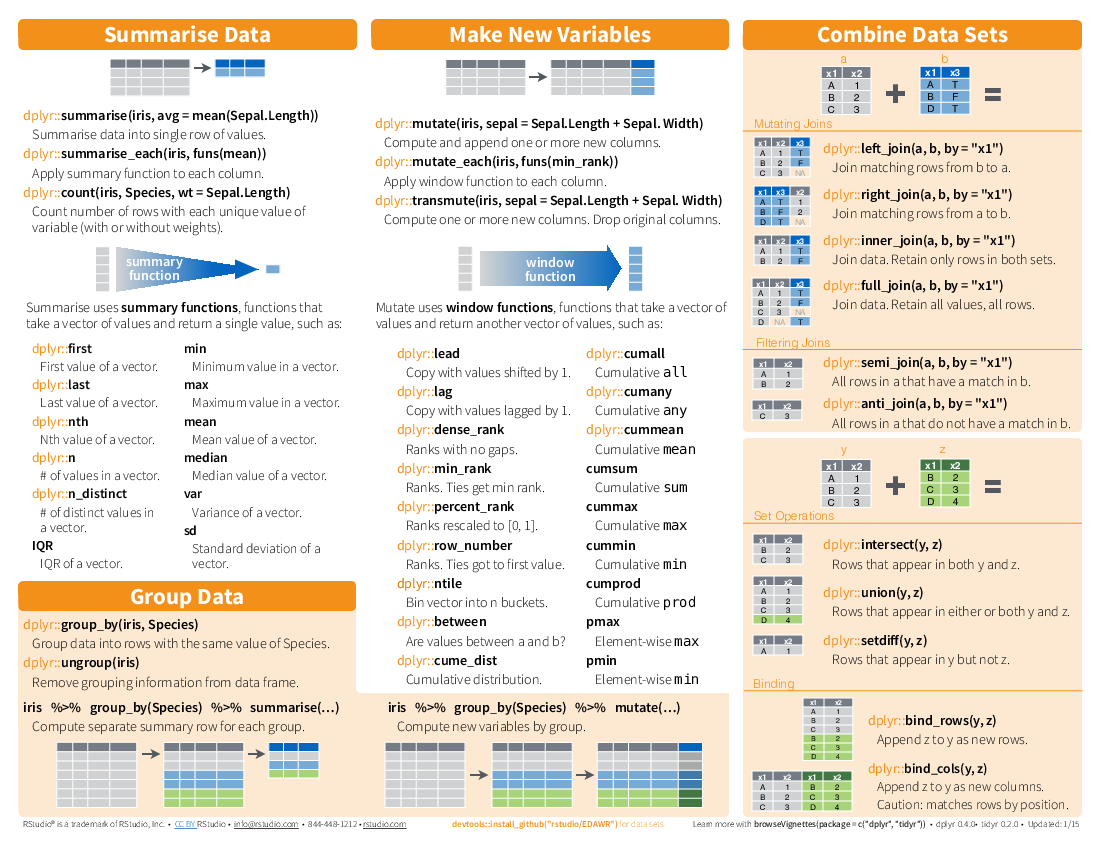
I have been using dplyr for a while and found it changing my codes in R scripts. It is very convenient as for data mungling and cleaning (whatever the verb you wish to use). The main syntax ‘%>%’ is almost like | in linux, any data frame can be passed/piped into the next verb.
library(dplyr)
## Warning: package 'dplyr' was built under R version 3.1.2
##
## Attaching package: 'dplyr'
##
## The following object is masked from 'package:stats':
##
## filter
##
## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, union
data(DNase)
head(DNase)
## Run conc density
## 1 1 0.04882812 0.017
## 2 1 0.04882812 0.018
## 3 1 0.19531250 0.121
## 4 1 0.19531250 0.124
## 5 1 0.39062500 0.206
## 6 1 0.39062500 0.215
summary(DNase)
## Run conc density
## 10 :16 Min. : 0.04883 Min. :0.0110
## 11 :16 1st Qu.: 0.34180 1st Qu.:0.1978
## 9 :16 Median : 1.17188 Median :0.5265
## 1 :16 Mean : 3.10669 Mean :0.7192
## 4 :16 3rd Qu.: 3.90625 3rd Qu.:1.1705
## 8 :16 Max. :12.50000 Max. :2.0030
## (Other):80
dat <- DNase %>%
filter(Run != 10) %>% # filtering any Run smaller than 10
tbl_df()
summary(dat)
## Run conc density
## 11 :16 Min. : 0.04883 Min. :0.0110
## 9 :16 1st Qu.: 0.34180 1st Qu.:0.1978
## 1 :16 Median : 1.17188 Median :0.5265
## 4 :16 Mean : 3.10669 Mean :0.7190
## 8 :16 3rd Qu.: 3.90625 3rd Qu.:1.1705
## 5 :16 Max. :12.50000 Max. :2.0030
## (Other):64
dat <- DNase %>%
mutate(Run_duplicate = Run) # make a new column with the verb 'mutate'
head(dat)
## Run conc density Run_duplicate
## 1 1 0.04882812 0.017 1
## 2 1 0.04882812 0.018 1
## 3 1 0.19531250 0.121 1
## 4 1 0.19531250 0.124 1
## 5 1 0.39062500 0.206 1
## 6 1 0.39062500 0.215 1
summaryTable <- dat %>%
group_by(Run) %>%
summarize(number_of_sample = n(), # group_by and summarize can be very useful for
sum_density = sum(density)) # getting summary statistics for different categories
summaryTable
## Source: local data frame [11 x 3]
##
## Run number_of_sample sum_density
## 1 10 16 11.530
## 2 11 16 11.358
## 3 9 16 11.294
## 4 1 16 10.833
## 5 4 16 10.890
## 6 8 16 11.292
## 7 5 16 11.274
## 8 7 16 11.728
## 9 6 16 11.937
## 10 2 16 12.044
## 11 3 16 12.392
from Rstudio blog